Performance
Performance
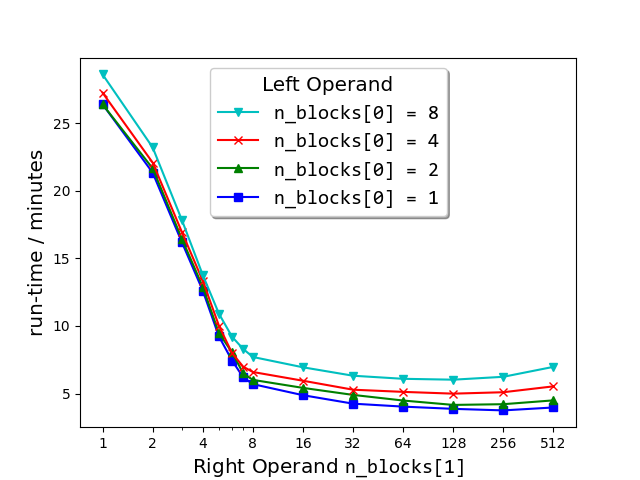
Semilogx plots of run-times of match_strings() vs the number of blocks (n_blocks[1]) into which the right matrix-operand of the dataset (663 000 strings from sec__edgar_company_info.csv) was split before performing the string comparison. As shown in the legend, each plot corresponds to the number n_blocks[0] of blocks into which the left matrix-operand was split.
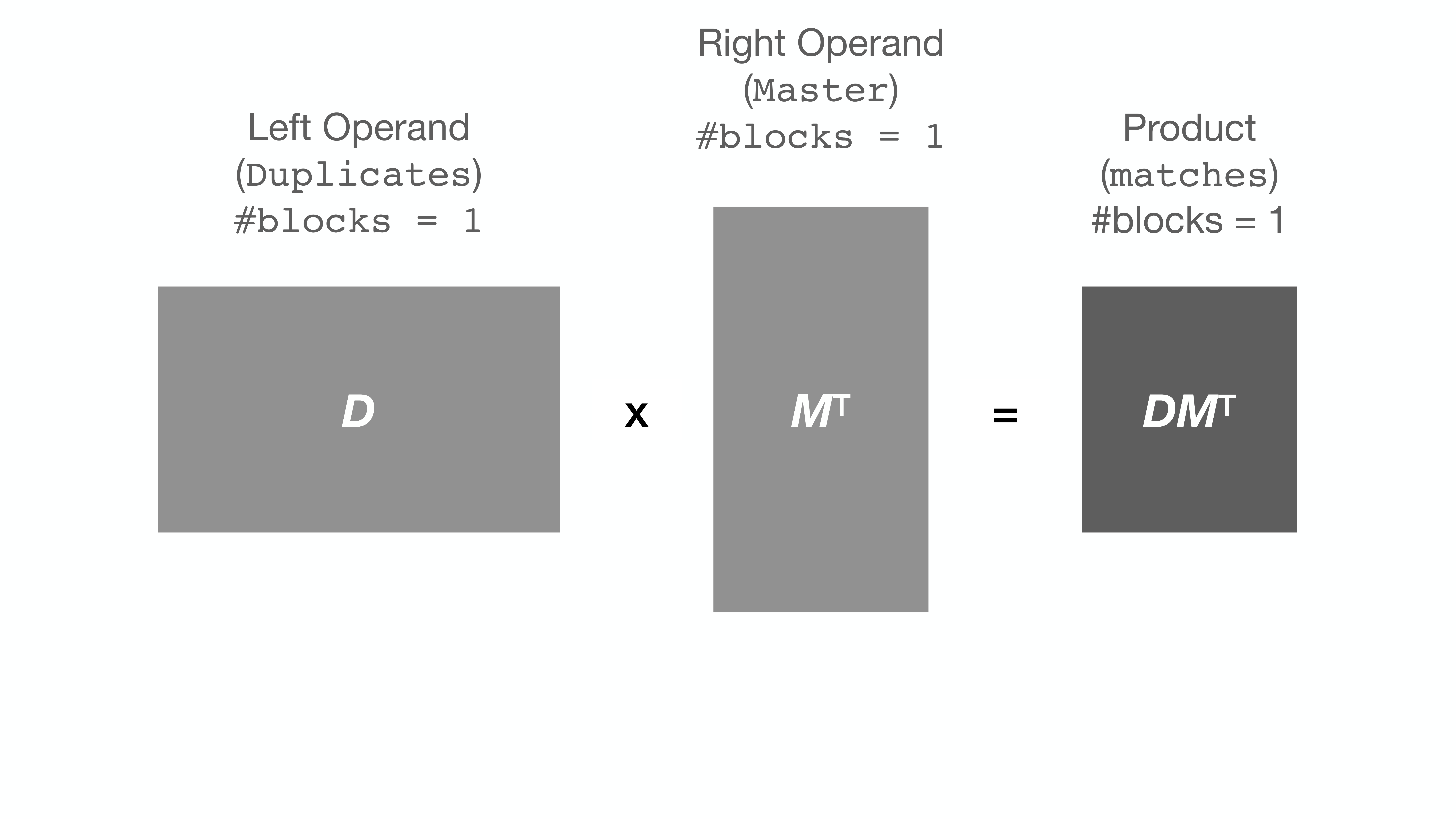
String comparison, as implemented by string_grouper, is essentially matrix
multiplication. A pandas Series of strings is converted (tokenized) into a
matrix. Then that matrix is multiplied by itself (or another) transposed.
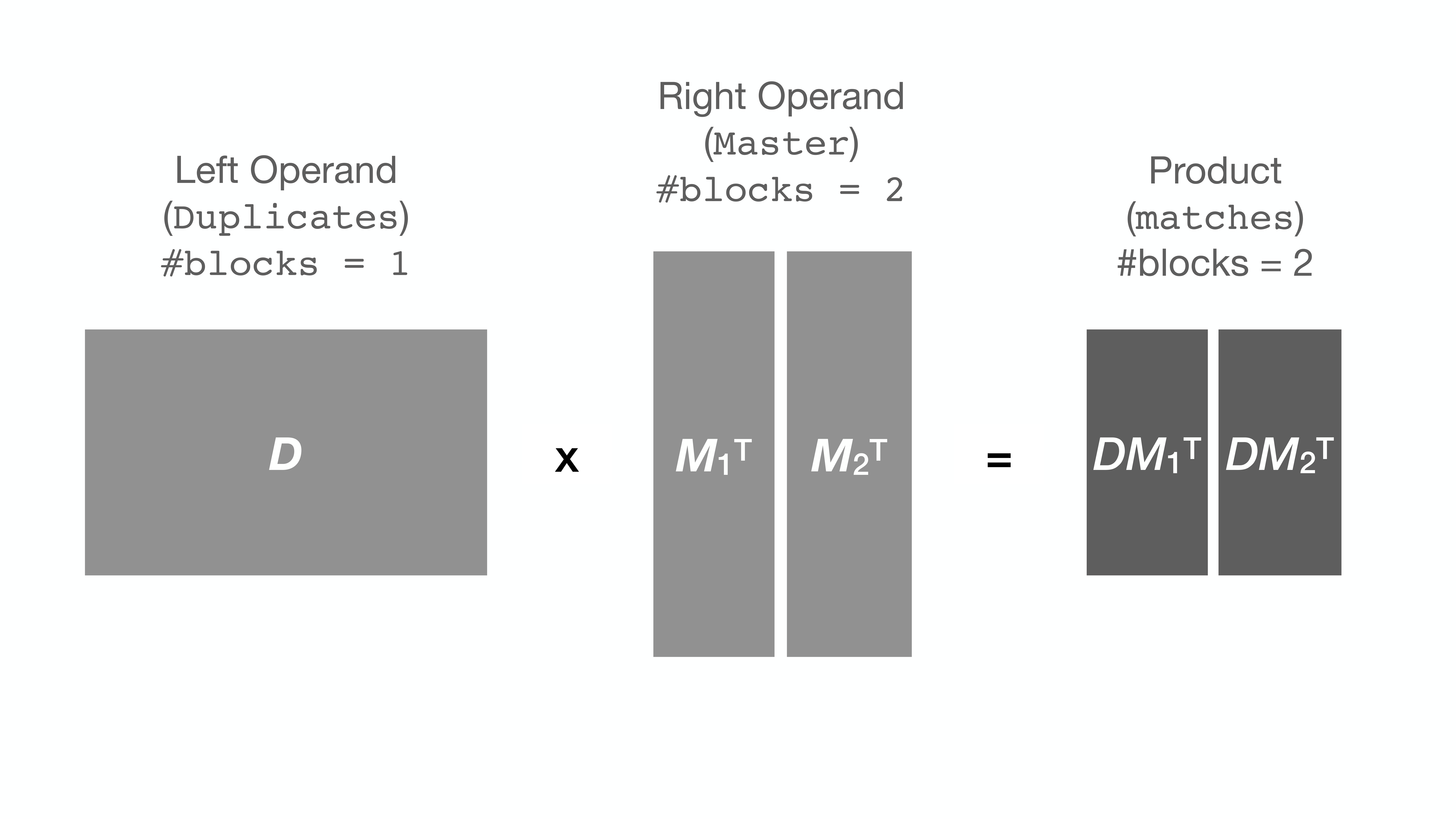
Here is an illustration of multiplication of two matrices D and MT:
It turns out that when the matrix (or Series) is very large, the computer
proceeds quite slowly with the multiplication (apparently due to the RAM being
too full). Some computers give up with an OverflowError.
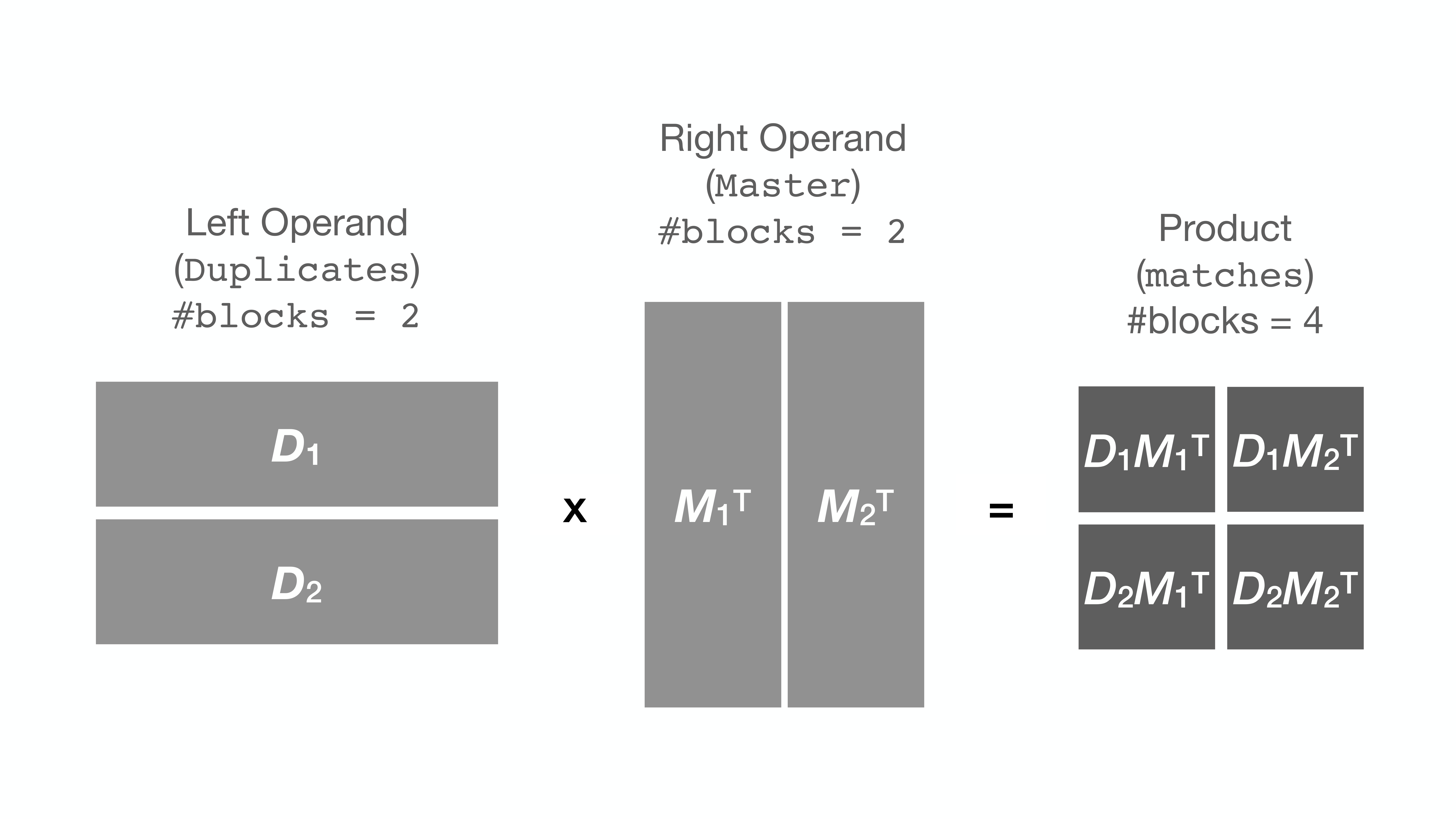
To circumvent this issue, string_grouper now allows the division of the Series
into smaller chunks (or blocks) and multiplies the chunks one pair at a time
instead to get the same result:
But surprise ... the run-time of the process is sometimes drastically reduced
as a result. For example, the speed-up of the following call is about 500%
(here, the Series is divided into 200 blocks on the right operand, that is,
1 block on the left × 200 on the right) compared to the same call with no
splitting [n_blocks=(1, 1), the default, which is what previous versions
(0.5.0 and earlier) of string_grouper did]:
# A DataFrame of 668 000 records:
companies = pd.read_csv('data/sec__edgar_company_info.csv')
# The following call is more than 6 times faster than earlier versions of
# match_strings() (that is, when n_blocks=(1, 1))!
match_strings(companies['Company Name')], n_blocks=(1, 200))
Further exploration of the block number space (see plot above) has revealed that for any fixed number of right blocks, the run-time gets longer the larger the number of left blocks specified. For this reason, it is recommended not to split the left matrix.
In general,
total runtime = n_blocks[0] × n_blocks[1] × mean runtime per block-pair
= Left Operand Size × Right Operand Size ×
mean runtime per block-pair / (Left Block Size × Right Block Size)
So for given left and right operands, minimizing the total runtime is the same as minimizing the
runtime per string-pair comparison ≝
mean runtime per block-pair / (Left Block Size × Right Block Size)
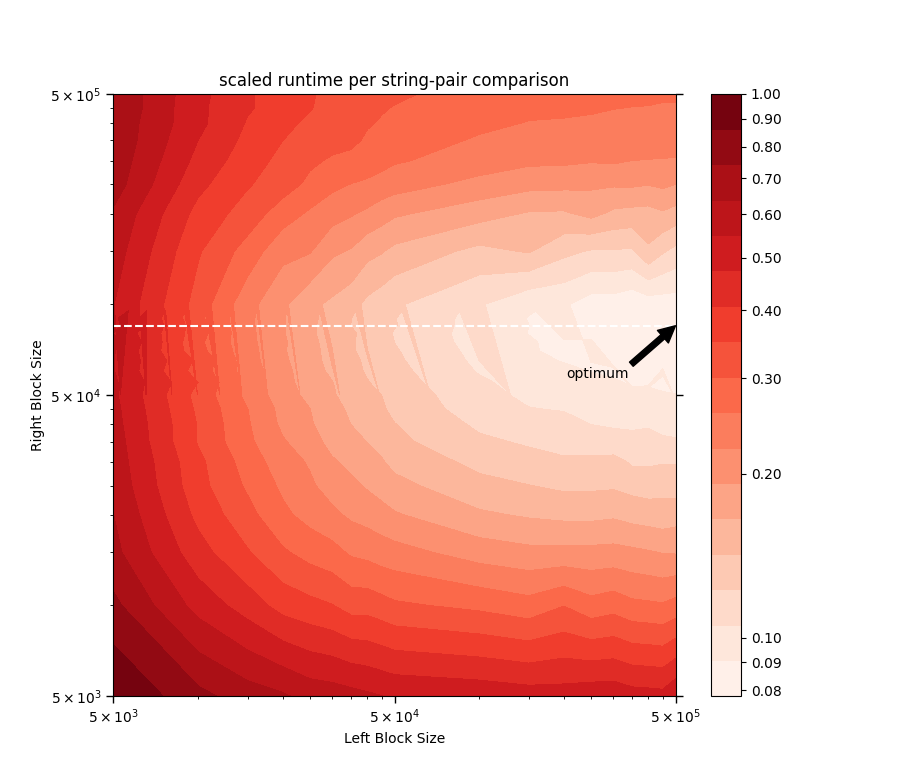
Below is a log-log-log contour plot of the runtime per string-pair comparison scaled by its value at Left Block Size = Right Block Size = 5000. Here, Block Size is the number of strings in that block, and mean runtime per block-pair is the time taken for the following call to run:
whereleft_Series and right_Series, corresponding to Left Block and Right Block respectively, are random subsets of the Series companies['Company Name')] from the
sec__edgar_company_info.csv sample data file.
It can be seen that when right_Series is roughly the size of 80 000 (denoted by the
white dashed line in the contour plot above), the runtime per string-pair comparison is at
its lowest for any fixed left_Series size. Above Right Block Size = 80 000, the
matrix-multiplication routine begins to feel the limits of the computer's
available memory space and thus its performance deteriorates, as evidenced by the increase
in runtime per string-pair comparison there (above the white dashed line). This knowledge
could serve as a guide for estimating the optimum block numbers —
namely those that divide the Series into blocks of size roughly equal to
80 000 for the right operand (or right_Series).
So what are the optimum block number values for any given Series? That is anyone's guess, and may likely depend on the data itself. Furthermore, as hinted above, the answer may vary from computer to computer.
We however encourage the user to make judicious use of the n_blocks
parameter to boost performance of string_grouper whenever possible.